DESIGNER-v2
Designer is a python tool for diffusion MRI preprocessing. It includes:
- denoising using MPPCA (or optionally using patch2self through dipy)
- RPG Gibbs artifact correction
- Rician bias correction
- EPI distortion correction
- Eddy current and motion correction
- b0 normalization for multiple input series
- b1 inhomogeneity correction
Designer was developed to simplify the often complex process of eliminating noise and artifacts from DICOM level diffusion data. It was written so that multiple input series can be preprocessed at once, in the case where some diffusion shells are acquired in separate series or acquisitions. Designer can support data with varying echo-time or b-shape depending on user input. It also supports complex-valued data, as users can import a diffusion phase map in order to eliminate the effects of the noise floor. It was originally developed by Benjamin Ades-Aron and Jenny Chen.
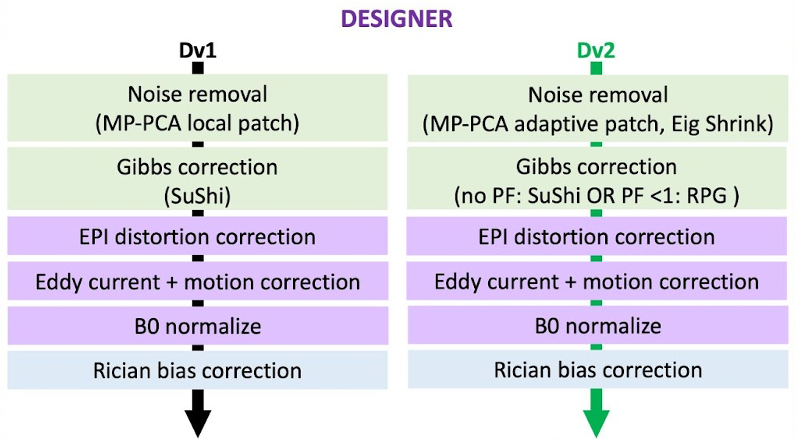 Designer version 2 has a number of notable changes from the original implementation. The majority of processing steps have been ported to either Python or C++ for speed, memory, and compatibility improvements. Beyond this, MPPCA and Gibbs correction techniques have been optimized. For details on these improvements please visit our background page.
Designer version 2 has a number of notable changes from the original implementation. The majority of processing steps have been ported to either Python or C++ for speed, memory, and compatibility improvements. Beyond this, MPPCA and Gibbs correction techniques have been optimized. For details on these improvements please visit our background page.
If you use designer please cite one of the following reference:
Ades-Aron, B., Veraart, J., Kochunov, P., McGuire, S., Sherman, P., Kellner, E., … & Fieremans, E. (2018). Evaluation of the accuracy and precision of the diffusion parameter EStImation with Gibbs and NoisE removal pipeline. Neuroimage, 183, 532-543.
Chen, J., Ades-Aron, B., Lee, H.H., Mehrin, S., Pang, M., Novikov, D. S., Veraart, J., & Fieremans, E. (2024). Optimization and validation of the DESIGNER preprocessing pipeline for clinical diffusion MRI in white matter aging. Imaging Neuroscience, 2 1–17
Tournier, J.-D.; Smith, R. E.; Raffelt, D.; Tabbara, R.; Dhollander, T.; Pietsch, M.; Christiaens, D.; Jeurissen, B.; Yeh, C.-H. & Connelly, A. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. NeuroImage, 2019, 202, 116137